Phenylalanine pairs capable of forming a molecular switch are also present in many other signaling proteins, including receptors in human cells, making it an attractive target for drug design and biotechnology applications.
However, using Anton, researchers were able to simulate only a small part of the chemoreceptor containing the Phe396 pair known as a dimer, meaning two identical molecules. But these two molecules do not work alone.
Dimers are grouped in threes to form larger units of the signaling complex, called trimers of dimers. Researchers expect simulating a trimer next will reveal more about how the Phe396-mediated signal is amplified across neighboring proteins.
But a trimer simulation requires modeling almost 400,000 atoms with increasingly complex physics calculations as the system gets larger. To do so, the group needs a lot of computational capacity.
"Anton is an exceptional machine, but its hardware limitations won't permit the simulation of such a large system," Ortega said. "We need Titan."
Using Titan they ran a preliminary simulation to determine that they would need millions of processing hours on this petaflop machine capable of quadrillions of calculations per second. Titan's GPUs are highly parallel, hurtling through repetitive calculations such as those modeling the large system of atoms under a vast array of configurations in the trimer simulation.
"With Titan we will begin to see how the signal propagates across chemoreceptors," Zhulin said. "We think this will start to explain how signals are amplified by these remarkable molecular machines."
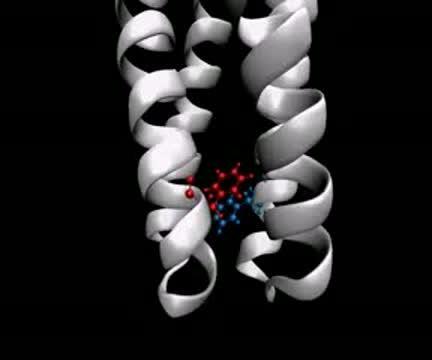
Watch the flipping conformations in a pair of phenylalanine amino acids called Phe396 (blue and red molecules), which act as a molecular switch essential to the signaling mechanism of an E. coli chemoreceptor.
(Photo Credit: Davi Ortega)
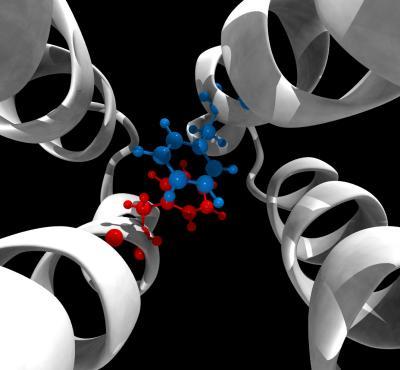
Red and blue molecules represent a conformational switch essential to the signaling mechanism of an E. coli chemoreceptor that researchers discovered using computational molecular dynamics simulations.
(Photo Credit: Davi Ortega)