X-ray crystallography reveals the three-dimensional structure of a molecule, thus making it possible to understand how it works and potentially use this knowledge to subsequently modulate its activity, especially for therapeutic or biotechnological purposes. For the first time, a study has shown that residual movements continue to animate proteins inside a crystal and that this movement "blurs" the structures obtained via crystallography. The study stresses that the more these residual movements are restricted, the better the crystalline order. That is why molecules consisting of the most compact crystals generally make it possible to obtain structures of better quality. This research combines crystallography, nuclear magnetic resonance (NMR) and simulation and is the result of an international cooperation involving researchers from the Institute of Structural Biology (ISB, CEA/CNRS/ Joseph Fourier University) in Grenoble, France, Purdue University, USA, and the Institute of Complex Systems (ICS-6: Structural Biochemistry) at Forschungszentrum Jülich in Germany. The results were published in Nature Communications.
X-ray crystallography is the most prolific method for determining protein structures. The quality of a crystallographic structure depends on the "degree of order" within the crystal. Proteins are generally only a few nanometres in size. Several thousand billion protein molecules must perfectly fit together in order to create a well-ordered crystalline structure in three dimensions. This stage is necessary if a structure is to be successfully obtained. Sometimes crystals, which may appear macroscopically perfect, disintegrate if subjected to X-rays, thus destroying their structure. It has been suggested that mass movements of crystalline proteins might explain this paradox, but this supposedly slow residual dynamic had never been observed directly in a crystal.
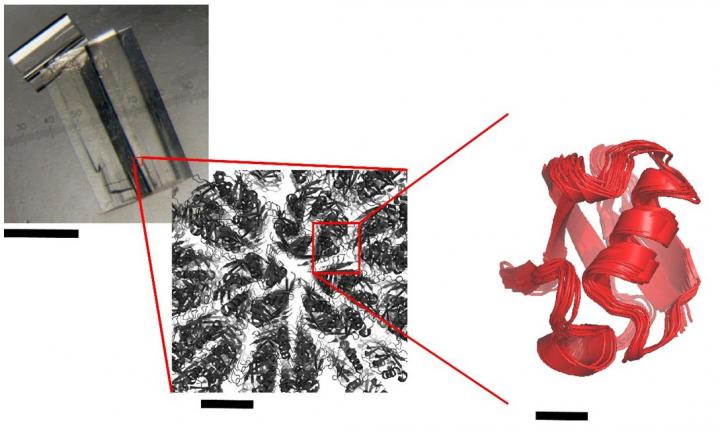
The researchers at IBS used a multi-technique approach, combining solid-state NMR spectroscopy, simulations of molecular dynamics and X-ray crystallography. Thanks to solid-state NMR, they were able to measure the dynamics of a model protein, ubiquitin, in three of its crystalline forms. Their results indicate that, even when crystallised, proteins continue to produce slight residual movements. The less compact the crystal, the more unrestrained the movements within it.
 Observation of a typical protein crystal used to study molecular structures by crystallography. Credit: Paul Schanda/CEA)
Observation of a typical protein crystal used to study molecular structures by crystallography. Credit: Paul Schanda/CEA)
Source: Paul Schanda/CEA