In the accompanying study, the researchers used data from the 1000 Genomes Project to analyze DNA sequence variations across the entire genome. They identified hundreds of gene variants that potentially contributed to human evolutionary adaptation. One of these variants, a mutation in the TLR5 gene, changed the immune responses of cells exposed to bacterial proteins, suggesting that this variant could confer a fitness advantage by protecting against bacterial infections. The comprehensive list of possible adaptive mutations driving recent human evolution provides the groundwork for future studies.
Together, the two studies represent a decisive shift for the field of evolutionary genomics, transitioning from hypothesis-driven to hypothesis-generating science. By moving from genome-wide scans to the characterization of adaptive mutations, it is possible to elucidate distinct mechanisms of evolution.
"These two studies are the product of work done in this area for over a decade but can only now be made possible with the major breakthroughs in genomic technology," Sabeti says. "I am struck by the ability of genomics to uncover the secrets of human history."

A pair of studies published by Cell Press on February 14th in the journal Cell sheds new light on genetic variation that may have played a key role in human evolution. The study researchers used an animal model to study a gene variant that could have helped humans adapt to humid climates, and they used whole-genome sequence data to identify hundreds of gene variants that potentially helped humans adapt to changing environmental conditions over time. The findings provide a road map for understanding human biological history as well as modern-day variability.
(Photo Credit: Cell, Grossman et al. and Kamberov et al.)
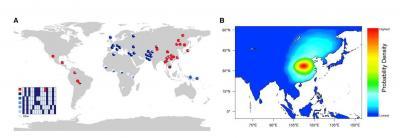
(A) This shows haplotype distribution of the genomic region surrounding V370A, based on 24 SNPs covering _139 kb. The six most common haplotypes are shown, and the remaining low-frequency haplotypes grouped as Other. The chimpanzee allele was assumed to be ancestral. Derived alleles are in dark blue, except for the 370A variant which is red.(B) The approximate posterior probability density for the geographic origin of 370A obtained by ABC simulation. The heat map was generated using 2D kernel density estimation of the latitude and longitude coordinates from the top 5,000 (0.46%) of 1,083,966 simulations. Red color represents the highest probability, and blue the lowest.
(Photo Credit: Cell, Kamberov et al. Figure 1)
Source: Cell Press