A noninvasive, sequencing-based approach for detecting chromosomal abnormalities in the developing fetus is safer and more informative in some cases than traditional methods, according to a study published by Cell Press January 10th in The American Journal of Human Genetics. This method, which analyzes fetal DNA in the mother's blood, could provide women with a cost-effective way to find out whether their unborn baby will have major developmental problems without risking a miscarriage.
"Our study is the first to show that almost all the information that is available from an invasive procedure is also available noninvasively from a simple maternal blood draw," says senior study author Richard Rava of Verinata Health.
Metaphase karyotypes—pictures of chromosomes taken through the microscope—have traditionally been used to detect abnormalities associated with developmental delay, intellectual disability, congenital defects, and autism. Recently, a method called a chromosome microarray, which uses molecular probes to detect gains or losses of DNA segments within chromosomes, has been shown to provide more detailed information than a metaphase karyotype. But both of these approaches require invasive procedures that involve removing tissue from the placenta or inserting a needle into the amniotic sac to collect fluid. As a result, they increase the risk of infection and could harm the fetus during pregnancy.
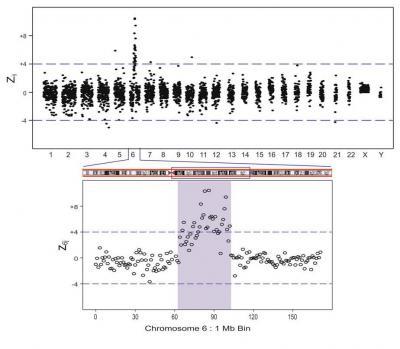
The expanded regions show z6j 1 Mb bin results. The figure shows a 38 Mb duplication, covering the region between 64 Mb and 102 Mb of Chr 6.
(Photo Credit: American Journal of Human Genetics, Srinivasan et al. Figure 3.)
Massively parallel sequencing (MPS) of fetal DNA in the mother's blood offers a safer alternative for the detection of chromosomal abnormalities across the fetal genome. But it has not been known whether MPS is as accurate as chromosome microarrays. In the new study, Rava and his team found that MPS was capable of detecting a variety of chromosomal abnormalities as accurately as chromosome microarrays.
"Such a noninvasive test could have clinical utility in the near future, particularly for women who either have a medical contraindication or lack access to an invasive procedure," Rava says. "This work suggests an exciting future path toward routine noninvasive detection of abnormalities in the entire fetal genome."
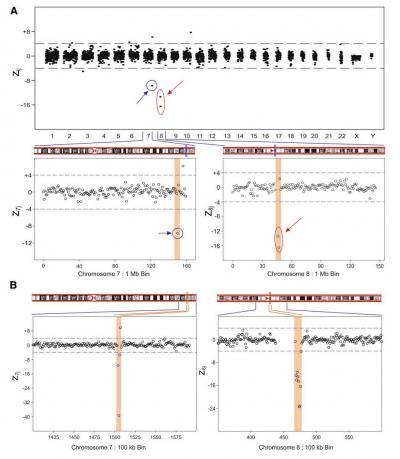
This clinical sample has a karyotype with a small deletion in chromosome 7 (blue circle). Another small deletion is detected in chromosome 8 (red circle). Expanded regions show z7j and z8j 1 Mb and 100 kb bin data. The figure shows a 1 Mb deletionat bin number 150 Mb on Chr 7 (A). At higher resolution (B), this deletion is found to be 300 kb long, in the region from 150.3 Mb to 150.6 Mb of Chr 7. Note: The putative copy-number gain seen in Chr 7 at bin number 156 in the 1 Mb data is notseen in the same region in the 100 kb data. The figure also shows a 2 Mb deletion on Chr 8 (A) covering bins 46 Mb and 47 Mb. At higher resolution (B), this resolves into a 900 kb deletion, covering the region from 46.9 Mb to 47.7 Mb of Chr 8.
(Photo Credit: American Journal of Human Genetics, Srinivasan et al. Figure 4.)
Source: Cell Press